In the framework of uLearnBio ICML 2014 workshop, we pppose three challenges on unsupervised learning from bioacoustic data : the first two ones concern both bird species clustering and unsupervised bird song decomposition and the third one concerns whale song decomposition. For this, we provide two challenging data sets : the birds data and the whale data.
Bird Challenge 1 Bird Challenge 2 Whale Challenge
Bird Challenge(1)
![[bird]](./images/a_bird.jpg)
The goal of the bird challenge is to build unsupervised learning models/techniques that can automatically infer the structure of bird song signals recorded for a given species (intra-species models), as well as to automatically retrieve species given several bird song records covering several species (inter-species models). These tasks are achieved from noisy complex data and can be performed using unsupervised techniques, in particular clustering, segmentation, topographic projection etc. In what follows we give a description of the provided data and what can expect as results by running unsupervised learning techniques on such data.
The data for this challenge are provided by Jerome Sueur from the Musee National d'Histoire Naturelle.
You find below the data sets. If you participate to this challenge, please let us know by email on uLearnBio@gmail.com for free inscription, and to get updated news if any on these data during the challenge. The subject of your email should contain "[uLearnBio bird challenge submission] ..."
Tasks description : for each test file, you have to either index the 35 bird species given the data set below (species clustering) or for a chosen species, decompose its song in "syllables" using unsupervised techniques. You can also perform the two at once for example by using an hierarchical model. All the approaches have to perform in an unsupervised way.
Submission : The challenge submission shall be sent to
uLearnBio@gmail.com in the format of a .pdf file (please if possible use the ICML 2014 format guidelines). The submission must describe the model(s)/approach(es) and contain the results by mentioning at least the following information details : the obtained structure (e.g., clustering partition into species on song decomposition/segmentation for a given species, etc), the evaluation criteria to judge the quality of the obtained partition (in case of clustering one can use for example the rand index, the silhouette coefficient, the intra/intra class inertia, etc).
For the bird challenge of species clustering, the submitted results can also b presented as Pij (90xN) probabilities, where N is the number of models of birds or other acoustic events, and Pij = P('The acoustic_event j occurs in the test file i').
Evaluations will take into account namely the quality of the partition (by the metrics given above), the capability is infer automatically the number of clusters/segments from the data in particular in the case where this one is assumed to be unknown for the approach (in bird species clustering as well as in bird song decomposition).
In case you prefer using your own features extracted from the .wav files, your submission may include if possible at least one result using the given MFCC features.
link to the data : 90 .wav files and suggested features (Mel Filter Cepstrum Coefficients (MFCC))
The suggested MFCC features were computed according to a minimum error (in average on all the species) reconstruction signal of the signal.
METADATA : You may also use the Geographical position of the 3 microphones, the Meteorological conditions during the recording and the Phylogenetic tree of the 35 possibly singing species in this site. It is forbidden to use any additional external data or recordings (as wikipedia wav samples nor other informations from ICML4B 2013. Do not use the training set from ICML 2013 workshop).
PHYLOGENETIC tree to reveal some acoustic cues between species. This tree, with distances, is given there.
The phylogenetic tree, with distances, of the target species.
WEATHER : The weather, wind speed, humidity, sun conditions... of each test set files.
You are invited to send a working note with your (partial) results by the 30th of may following the ICML 2014 template: it will be published into this ICML workshop Proceedings.
Bird Challenge (2): LifeClef Bird challenge
![[birdBrazil]](./images/BirdBrazilMini.jpg)
The second subtask of this challenge will be based on the same (training) dataset of the LifeCLEF 2014 Bird task (http://www.imageclef.org/2014/lifeclef/bird), a parallel event on
bird identification. The main originality of this data is that it was specifically built through a citizen sciences initiative conducted by Xeno-canto (http://www.xeno-canto.org/), an international social network of amateur and expert ornithologists. The dataset contains
around 10k audio records on 500 species from Brazil.
Please use the LifeCLEF/ImageCLEF registration interface (http://medgift.hevs.ch:8080/CLEF2014/faces/Login.jsp) and subscribe to the « LifeCLEF2014:bird » substrack in order to get the download link to the dataset. If you already have a login from the former ImageCLEF benchmarks you can migrate it to LifeCLEF 2014 here (http://medgift.hevs.ch:8080/ImageClefEvents/).
In order to allow metrics and comparisons, please use this TRAIN / TEST partitions as defined in this .xls with the species label and the file location, date... useful to analyse your clusters (updated 13th march)
: SET_DEFINITIONS and LABELS.
Baseline .MFCC features designed for bird song analysis in NIPS 2013
(16 coeff + delta and deltadelta), and used in the supervised task in
LifeClef, are also available for this challenge for the two data sets
in the same repertory than the .wav in LifeClef Bird Challenge web
site. You might use them for a fast prototype.
This large bird challenge is related to 500 amazonian species. The goal is to retrieve / classify into an unsupervised way and to show on real material how unsupervised learning techniques can namely help to structure and “label” rare bird species and/or birds songs.
Notice that the labels of all the recordings are available for this task, however the proposed techniques should perform in unsupervised way so that the labels will be used only for performance evaluation, with your metrics or standard metrics on clustering partitions evaluation (e.g., misclassification error rate, the rand index, the silhouette coefficient, the intra/intra class inertia, etc).
If you participate to one of these challenges, please also let us know by email on uLearnBio@gmail.com
for free inscription, and to get updated news if any on these data during the challenge. The subject of your email should contain "[uLearnBio LifeClef bird challenge submission] ..."
Bird_ULB challenge organizers : H. Glotin UTLN, H. Goëau, INIRIA, A. Rauber, TU Wien, W. Vellinga, Xeno-Canto, F. Chamroukhi, UTLN
Whale Challenge

The goal of the whale challenge is to build unsupervised learning models/techniques that can automatically infer the structure of humpback whale song. Humpback whales produce songs with a specific structure and the study of that songs is very challenging and very useful for bio-acousticians and scientists to namely understand how do whales song, that is according to which "vocabulary". The problem can then be seen as the one of discovering the call units. This analysis can be seen as a problem of unsupervised call units discovery and can also be performed using unsupervised techniques, namely particular clustering, segmentation, etc.
It has been well documented that Humpback whales produce songs with a specific structure. We provide 26 minutes of a remarkable Humpback whale song recording produced at few meters distance from the whale in La Reunion - Indian Ocean, by our "Darewin" research group in 2013 (frequency sample = 44.1kHz, 32 bits, mono, wav, 130MB). Upload here 26 minutes of a remarkable Humpback whale song recording.
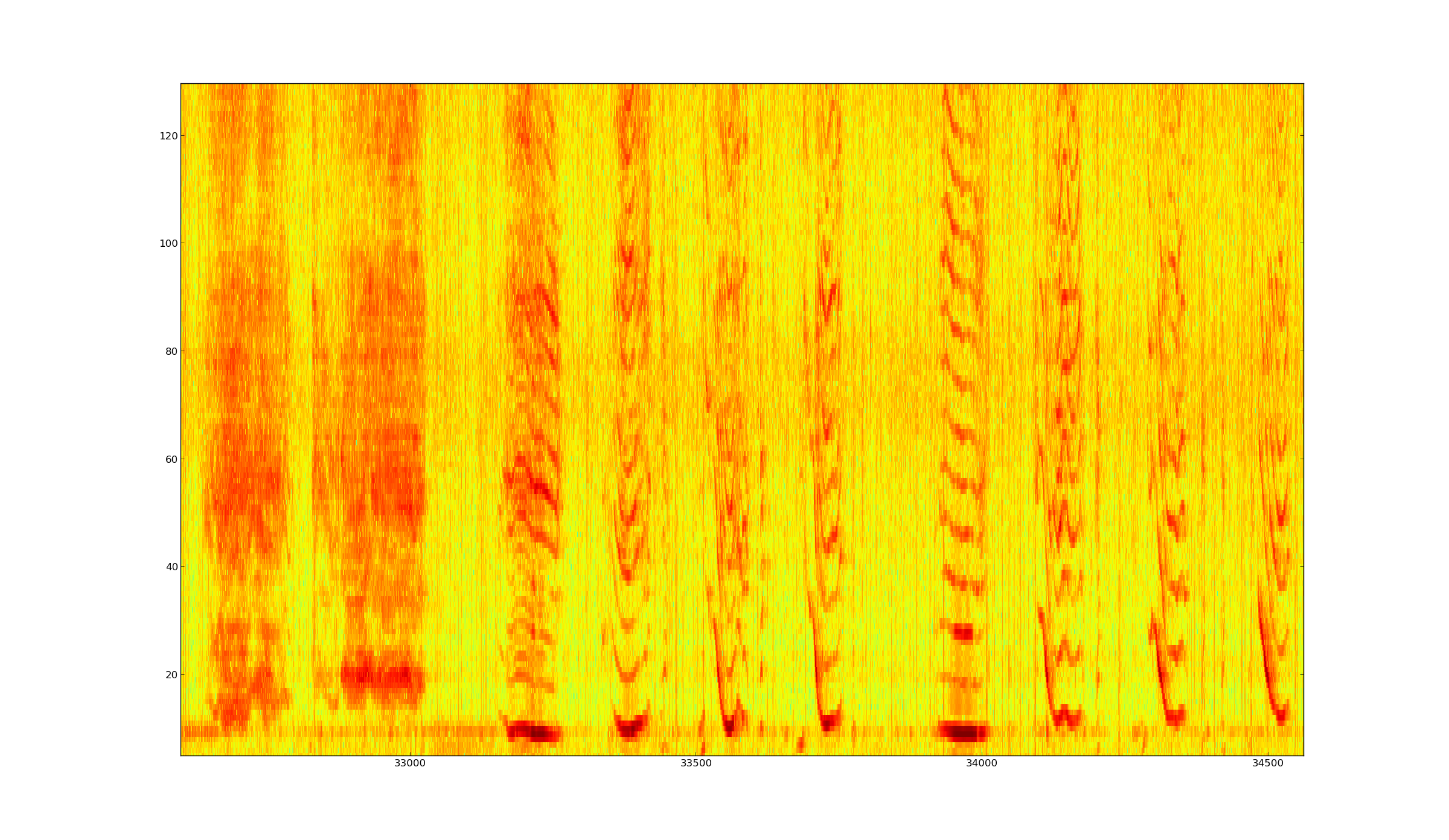
Figure: Spectrum of around 20 seconds of the given song of Humpback Whale (start from about 5'40 to 6'. Ordinata from 0 to 22.05 kHz, over 512 bins (fft on 1024 bins), frameshift of 10 ms.
You can download the Mel Filter Cepstrum Coefficients (MFCC) of this wav file (octave / matlab v6 format). The parameters of extraction of these MFCC are given here..
For this challenge, you may propose any efficient representation of this song that helps to study its structure, discover and index its song units. You can find an interesting linked approaches in:
- Pace, F., Benard, F., Glotin, H., Adam, O., and White, P. (2010) Subunit definition for humpback whale call classification, int. journal Applied Acoustics, Elsevier, 11(71):1107–1112
- Roch MA, Soldevilla MS, Burtenshaw JC, Henderson EE, Hildebrand JA (2007) Gaussian mixture model classification of odontocetes in the Southern California Bight and the Gulf of California. J. Acoust. Soc. America 121(3):1737-48.
The workshop will allow discussions over the proposed representations (clustering, indexing, sequence modeling etc.). The description of your approach (pdf) and your representation/results for this song file (.xml, .csv or .mat ...) shall be sent to uLearnBio@gmail.com. You shall include evaluation criteria to judge the quality of the obtained decomposition (in case of clustering one can use for example the rand index, the silhouette coefficient, the intra/intra class inertia, etc).
If you participate to this challenge, please let us know by email on uLearnBio@gmail.com for free inscription, and to get updated news if any on these data during the challenge. The subject of your email should contain "[uLearnBio whale challenge submission] ..."
Important dates
- The Challenge keynote deadline is the 30 may (Extended deadline)
- Final versions due (paper and challenge keynote) : 15 june 2014
- Note that the deadline for regular paper not related to the challenge is ealier: 25th april (extended deadline), with author notification the 30th april.

